Hydrosensing has published a new article “Construction of multi-targeted CRISPR libraries in tomato to overcome functional redundancy at genome-scale levels” in Nature Communications.
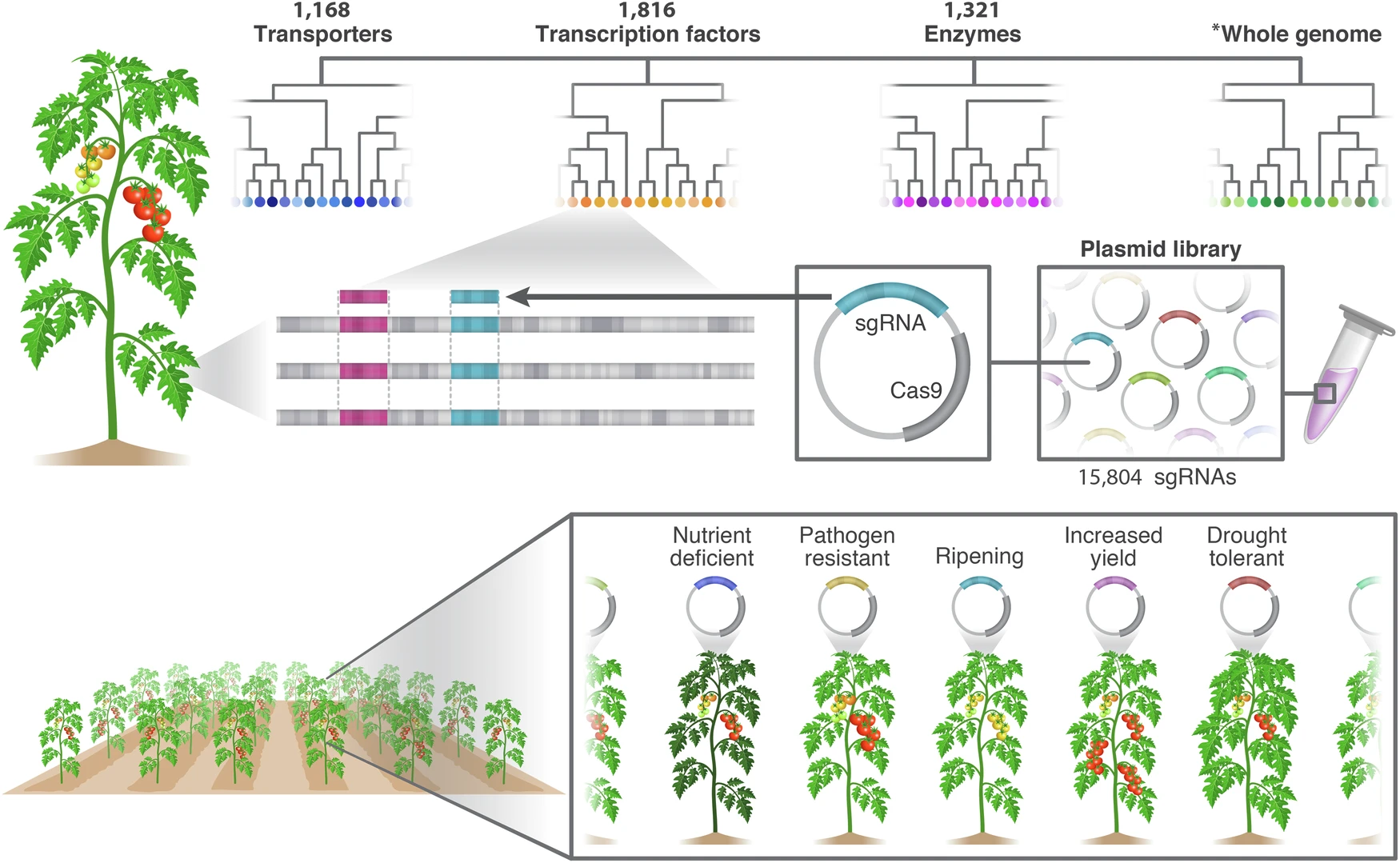
Genetic diversity is key for crop breeding and studying mutant traits, but traditional mutagenesis struggles with two big hurdles: overlapping gene functions and imprecise targeting. CRISPR-Cas9 solves the precision problem by editing genes at specific locations, but using it on a large scale in crops has been tough. In this study, we created a genome-wide, multi-target CRISPR library in tomato to scale up gene editing while keeping its pinpoint accuracy. We designed 15,804 unique guide RNAs (sgRNAs), each hitting several genes within the same gene family, and grouped them into 10 sub-libraries by gene function. From this, we produced about 1,300 distinct CRISPR tomato lines and uncovered mutants with clear traits linked to fruit development, flavor, nutrient absorption, and disease resistance. To track this at scale, we built CRISPR-GuideMap—a dual-barcode system for following sgRNAs in plants. Our findings show that multi-target CRISPR libraries can work at large scale, making them powerful tools for cracking gene redundancy and advancing both crop research and breeding.
Berman, A., Su, N., Li, Z. et al. Construction of multi-targeted CRISPR libraries in tomato to overcome functional redundancy at genome-scale level. Nat Commun 16, 4111 (2025). https://doi.org/10.1038/s41467-025-59280-6
Image: Schematic overview illustrating the library workflow, from design to screening. All coding genes were divided into phylogenetic trees, and trees were classified by function. sgRNAs were designed to target multiple genes located in close proximity to one another phylogenetically (indicated by colors). Each sgRNA was cloned into the Cas9 vector, creating a plasmid library. Transformed lines were screened with multi-targeted, large-scale, forward genetics for specific traits of interest, revealing hidden phenotypes. *Whole genome indicates all coding genes excluding transporters, transcription factors, and enzymes. Credit: Nat Commun