Hydrosensing Project
Discovering How Plants Sense Water Stress
The Hydrosensing Project Team is on a mission to transform our understandingof how plants sense and respond to water availability. We aim to uncover the mechanisms plants use to perceive water stress, a key factor in their survival and productivity.
By combining cutting-edge genomics, structural biology, biophysics and imaging approaches, we strive to revolutionize crop resilience and pave the way for climate-proof agriculture.
Join us as we explore new frontiers in plant science, working towards a future where crops are better equipped to withstand the challenges of a changing climate.
Publications
Journal articles and preprints by the Hydrosensing project
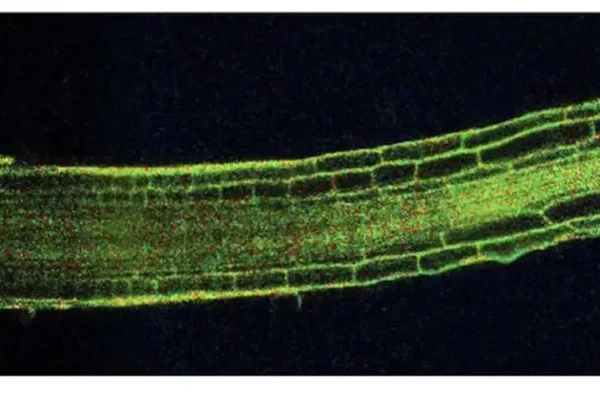
The mechanical properties of Arabidopsis thaliana roots adapt dynamically during development and to stress
Luis Alonso Baez et al., The mechanical properties of Arabidopsis thaliana roots adapt dynamically during development and to stress. Sci. Adv. 12,eaeb0032(2026).DOI:10.1126/sciadv.aeb0032

Transgenerational polarity axis inheritance during Ceratopterisembryogenesis
Woudenberg, S., Plackett, A. R., Hao, Z., Suzuki, H., Baez, L. A., Borassi, C., ... & Weijers, D. (2025). Transgenerational polarity axis inheritance during Ceratopteris embryogenesis. bioRxiv, 2025-08 https://doi.org/10.1101/2025.08.29.673061
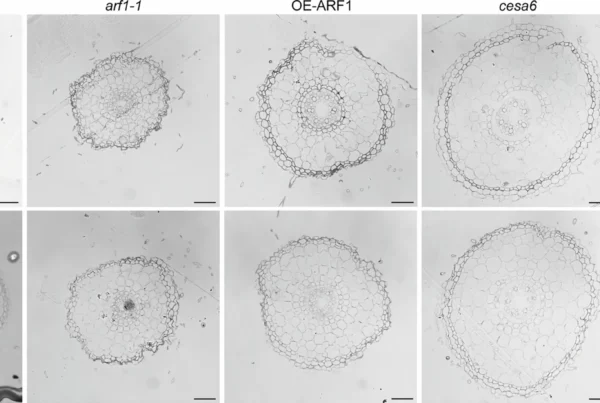
Ethylene modulates cell wall mechanics for root responses to compaction
Zhang, J., Liu, Z., Farrar, E.J. et al. Ethylene modulates cell wall mechanics for root responses to compaction. Nature 649, 685–692 (2026). https://doi.org/10.1038/s41586-025-09765-7

Developmental pathways in plants: Lessons from Arabidopsis for crop innovation
Malcolm J Bennett, Rahul Bhosale, Scott A Boden, Shu-Yan Chen, Tino Colombi, Toshiro Ito, Hong-Ju Li, Poonam Mehra, Lars Østergaard, Meng Li, Liu Liu, Nana Otsuka, Bipin K Pandey, Scott Poethig, Kalika Prasad, Yue Qu, Makoto Shirakawa, Ying Hua Su, Cao Xu, Wei-Cai Yang, Wen Jie Zhang, Xiaolan Zhang, Xian…


